Specifying Study Parameters
Maestro supports parameterization as a means of iterating over steps in a study with varying information. Maestro uses token replacement to define variables in the study specification to be replaced when executing the study. Token replacement can be used in various contexts in Maestro; however Maestro implements specific features for managing parameters.
Maestro makes no assumptions about how parameters are defined or used in a study, enabling flexibility in regards to how a study may be configured with parameters.
There are two ways Maestro supports parameters:
-
Directly in the study specification as the
global.parametersblock -
Through the use of a user created Python function called pgen
Maestro Parameter Block
The quickest and easiest way to setup parameters in a Maestro study is by defining a global.parameters block directly in the specification
global.parameters:
TRIAL:
values : [1, 2, 3, 4, 5, 6, 7, 8, 9]
label : TRIAL.%%
SIZE:
values : [10, 10, 10, 20, 20, 20, 30, 30, 30]
label : SIZE.%%
ITERATIONS:
values : [10, 20, 30, 10, 20, 30, 10, 20, 30]
label : ITER.%%
The above example defines the parameters TRIAL, SIZE, and ITERATIONS. Parameters can be used in study steps to vary information. When a parameter is defined in a study, Maestro will automatically detect the usage of a parameter moniker and handle the substitution automatically in the study expansion. This ensures that each set of parameters are run as part of the study.
The label key in the block specifies the pattern to use for the directory name when the workspace is created. By default, Maestro constructs a unique workspace for each parameter combination.
Defining multiple parameters in the parameter block will share a 1:1 mapping. Maestro requires all combinations be resolved when using the parameter block. The combinations in the above example will be expanded as follows:
-
TRIAL.1.SIZE.10.ITERATIONS.10
-
TRIAL.2.SIZE.10.ITERATIONS.20
-
TRIAL.3.SIZE.10.ITERATIONS.30
-
...
Maestro does not do any additional operations on parameters such as Cartesian products. If more complex methodologies are required to define parameters then the use of Maestro's pgen is recommended.
Note
Even when using the pgen functionality from the command line, Maestro will still initially verify that the provided specification is valid as if it planned to use it entirely (without pgen). If you are using the global.parameters block solely as documentation, we recommend that you comment out the global.parameters block. This lets the validator ignore it.
Defined parameters can be used in steps directly:
- name: run-lulesh
description: Run LULESH.
run:
cmd: |
$(LULESH)/lulesh2.0 -s $(SIZE) -i $(ITERATIONS) -p > $(outfile)
depends: [make-lulesh]
Even though this is defined in Maestro as a single step, Maestro will automatically run this step with each parameter combinations. This makes it very easy to setup studies and apply parameters to be run.
Note
Maestro will only use parameters if they've been defined in at least one step
In addition to direct access to parameter values, a parameter label can be used in steps by appending the .label moniker to the name (as seen below with $(ITERATIONS.label)):
- name: run-lulesh
description: Run LULESH.
run:
cmd: |
echo "Running case: $(SIZE.label), $(ITERATIONS.label)"
$(LULESH)/lulesh2.0 -s $(SIZE) -i $(ITERATIONS) -p > $(outfile)
depends: [make-lulesh]
What can be Parameterized in Maestro?
A common use case for Maestro is to use the parameter block to specify values to iterate over for a simulation parameter study; however, Maestro does not make any assumptions about what these values are. This makes the use of Maestro's parameter block very flexible. For example, Maestro does not require the parameter variations to be numeric.
study:
- name: run-simulation
description: Run a simulation.
run:
cmd: |
$(CODE) -in $(SPECROOT)/$(INPUT)
depends: []
global.parameters:
INPUT:
values: [input1.in, input2.in, input3.in]
label: INPUT.%%
The above example highlights a partial study spec that defines a parameter block of simulation inputs that will be varied when the study runs. The run-simulation step will run three times, once for each defined input file.
study:
- name: run-simulation
description: Run a simulation.
run:
cmd: |
$(CODE_PATH)/$(VERSION)/code.exe -in $(SPECROOT)/$(INPUT)
depends: []
global.parameters:
INPUT:
values : [input1.in, input2.in, input3.in, input1.in, input2.in, input3.in]
label : INPUT.%%
VERSION:
values : [4.0.0, 4.0.0, 4.0.0, 5.0.0, 5.0.0, 5.0.0]
label : VERSION.%%
This example parameterizes the inputs and the version of the code being run. Maestro will run each input with the different code version. The above example assumes that all the code versions share a base path, $(CODE_PATH) which is inserted via the token replacment mechanism from the env block to yeild the full paths (e.g. /usr/gapps/code/4.0.0/code.exe).
Where can Parameters be used in Study Steps?
Maestro uses monikers to reference parameters in study steps, and will automatically perform token replacement on used parameters when the study is run. The page Maestro Token Replacement goes into detail about how token replacement works in Maestro.
Maestro is very flexible in the way it manages token replacement for parameters and as such tokens can be used in a variety of ways in a study.
Cmd block
Parameters can be defined in the Maestro cmd block in the study step. Everything in Maestro's cmd block will be written to a bash shell or batch script (if batch is configured). Any shell commands should be valid in the cmd block. A common way to use parameters is to pass them in via arguments to a code, script, or tool.
...
- name: run-simulation
description: Run a simulation.
run:
cmd: |
/usr/gapps/code/bin/code -in input.in -def param $(PARAM)
depends: []
...
The specific syntax for using a parameter with a specific code, script, or tool will depend on how the application supports command line arguments.
Batch Configuration
Step based batch configurations can also be parameterized in Maestro. This provides an easy way to configure scaling studies or to manage studies where batch settings are dependent on the parameter values.
study:
- name: run-simulation
description: Run a simulation.
run:
cmd: |
$(CODE_PATH)/$(VERSION)/code.exe -in input.in -def RES $(RES)
procs: $(PROC)
nodes: $(NODE)
walltime: $(WALLTIME)
depends: []
global.parameters:
RES:
values : [2, 4, 6, 8]
label : RES.%%
PROC:
values : [8, 8, 16, 32]
label : PROC.%%
NODE:
values : [1, 1, 2, 4]
label : NODE.%%
WALLTIME:
values : ["00:10:00", "00:15:00", "00:30:00", "01:00:00"]
label : PROC.%%
Parameter Generator (pgen)
Maestro's Parameter Generator (pgen) supports setting up more flexible and complex parameter generation. Maestro's pgen is a user supplied python file that contains the parameter generation logic, overriding the global.parameters block in the yaml specification file. To run a Maestro study using a parameter generator just pass in the path to the pgen file to Maestro on the command line when launching the study, such as this example where the study specification file and pgen file live in the same directory:
The minimum requirements for making a valid pgen file is to make a function called get_custom_generator, which returns a Maestro ParameterGenerator object as demonstrated in the simple example below:
from maestrowf.datastructures.core import ParameterGenerator
def get_custom_generator(env, **kwargs):
p_gen = ParameterGenerator()
params = {
"COUNT": {
"values": [i for i in range(1, 10)],
"label": "COUNT.%%"
},
}
for key, value in params.items():
p_gen.add_parameter(key, value["values"], value["label"])
return p_gen
The object simply builds the same nested key:value pairs seen in the global.parameters block available in the yaml specification.
For this simple example above, this may not offer compelling advantages over writing out the flattened list in the yaml specification directly. This programmatic approach becomes preferable when expanding studies to use hundreds of parameters and parameter values or requiring non-trivial parameter value distributions. The following examples will demonstrate these scenarios using both standard python library tools and additional 3rd party packages from the larger python ecosystem.
Example: Lulesh Itertools
Using Python's itertools package from the standard library to perform a Cartesian Product of parameters in the lulesh example specification.
This results in the following set of parameters, matching the lulesh sample workflow:
| Parameter | Values | ||||||||
| TRIAL | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| SIZE | 10 | 10 | 10 | 20 | 20 | 20 | 30 | 30 | 30 |
| ITER | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
flowchart TD;
A(study root) --> COMBO1;
subgraph COMBO1 [Combo #1]
subgraph run-simulation1 [run-simulation]
B("TRIAL: 0\n SIZE: 10\n ITER: 1")
end
end
style B text-align:justify
A --> COMBO2
subgraph COMBO2 [Combo #2]
subgraph run-simulation2 [run-simulation]
C("TRIAL: 1\n SIZE: 10\n ITER: 2")
end
end
style C text-align:justify
A --> COMBO3
subgraph COMBO3 [Combo #3]
subgraph run-simulation3 [run-simulation]
D("TRIAL: 2\n SIZE: 10\n ITER: 3")
end
end
style D text-align:justify
A --> COMBO4
subgraph COMBO4 [Combo #4]
subgraph run-simulation4 [run-simulation]
E("TRIAL: 3\n SIZE: 20\n ITER: 4")
end
end
style E text-align:justify
A --> COMBO5
subgraph COMBO5 [Combo #5]
subgraph run-simulation5 [run-simulation]
F("TRIAL: 4\n SIZE: 20\n ITER: 5")
end
end
style F text-align:justify
A --> COMBO6
subgraph COMBO6 [Combo #6]
subgraph run-simulation6 [run-simulation]
G("TRIAL: 5\n SIZE: 20\n ITER: 6")
end
end
style G text-align:justify
A --> COMBO7
subgraph COMBO7 [Combo #7]
subgraph run-simulation7 [run-simulation]
H("TRIAL: 6\n SIZE: 30\n ITER: 7")
end
end
style H text-align:justify
A --> COMBO8
subgraph COMBO8 [Combo #8]
subgraph run-simulation8 [run-simulation]
I("TRIAL: 7\n SIZE: 30\n ITER: 8")
end
end
style I text-align:justify
A --> COMBO9
subgraph COMBO9 [Combo #9]
subgraph run-simulation9 [run-simulation]
J("TRIAL: 8\n SIZE: 30\n ITER: 9")
end
end
style J text-align:justifyPgen Arguments (pargs)
There is an additional pgen feature that can be used to make them more dynamic. The above example generates a fixed set of parameters, requiring editing the lulesh_itertools_pgen file to change that. Maestro supports passing arguments to these generator functions on the command line:
$ maestro run study.yaml --pgen itertools_pgen_pargs.py --parg "SIZE_MIN:10" --parg "SIZE_STEP:10" --parg "NUM_SIZES:4"
Each argument is a string in key:value form, which can be accessed in the parameter generator function as shown below:
Passing the pargs `SIZE_MIN:10', 'SIZE_STEP:10', and 'NUM_SIZES:4' then yields the expanded parameter set:
| Parameter | Values | |||||||||||
| TRIAL | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
| SIZE | 10 | 10 | 10 | 20 | 20 | 20 | 30 | 30 | 30 | 40 | 40 | 40 |
| ITER | 10 | 20 | 30 | 10 | 20 | 30 | 10 | 20 | 30 | 10 | 20 | 30 |
Notice that using the pgen input method makes it trivially easy to add 1000's of parameters, something which would be cumbersome via manual editing of the global.parameters block in the study specification file.
There are no requirements to cram all of the logic into the get_custom_generator function. The next example demonstrates using 3rd party libraries and breaking out the actual parameter generation algorithm into separate helper functions that the get_custom_generator function uses to get some more complicated distributions. The only concerns with this approach will be to ensure the library is installed in the same virtual environment as the Maestro executable you are using. The simple parameter distribution demoed in here is often encountered in polynomial interpolation applications and is designed to suppress the Runge phenomena by sampling the function to be interpolated at the Chebyshev nodes.
Example: Parameterized Chebyshev Sampling with pargs
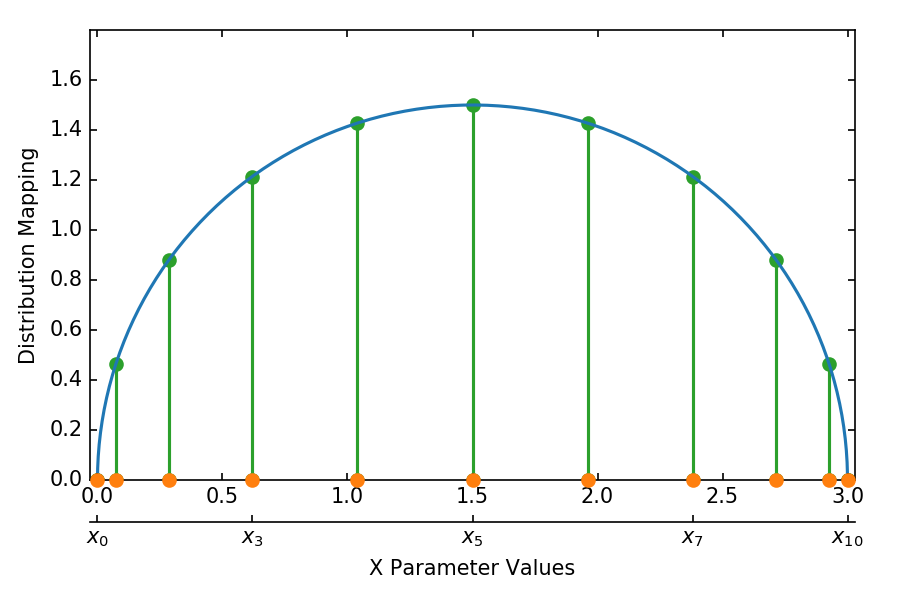
Using numpy to calculate a sampling of a function at the Chebyshev nodes.
Running this parameter generator with the following pargs
$ maestro run study.yaml --pgen np_cheb_pgen.py --parg "X_MIN:0" --parg "X_MAX:3" --parg "NUM_PTS:11"
results in the 1D distribution of points for the X parameter shown by the orange circles:
Referencing Values from a Specification's Env Block
In addition to command line arguments via pargs, the variables defined in the env block in the workflow specification file can be accessed inside the ParameterGenerator objects, which is passed in to the user defined get_custom_generator function as the first argument. The lulesh sample specification can be extended to store the default values for the pgen, enhancing the reproducability of the generator. The following example makes use of an optional seed parameter which can be added to the env block or set via pargs to make a repeatable study specification, while omitting it can enable fully randomized workflows upon every instantiation. The variables are accessed via the StudyEnvironment objects' find() function, which will return None if the variable is not defined in the study specification.